
Evidence based medicine is defined as using the best available evidence for everyday clinical practice [1–3]. Synthetic literature including systematic reviews and meta-analyses plays an important role in evidence based medicine. Essentially systematic reviews and meta-analyses are the cornerstone of evidence based practice. The main difference between a systematic review and a narrative review is the clear method of the former including a clear search and predefined inclusion criteria. The methodology of systematic reviews makes them reproducible which is not the case in narrative reviews [1–3]. The number of systematic reviews and meta-analyses on nuclear medicine diagnostic and prognostic studies is increasing [4, 5]. In the current chapter, a practical guideline has been prepared for the researchers who intend to perform a systematic review or meta-analysis of diagnostic and prognostic studies.
You have full access to this open access chapter, Download chapter PDF

Evidence based medicine is defined as using the best available evidence for everyday clinical practice [1,2,3]. Synthetic literature including systematic reviews and meta-analyses plays an important role in evidence based medicine. Essentially systematic reviews and meta-analyses are the cornerstone of evidence based practice. The main difference between a systematic review and a narrative review is the clear method of the former including a clear search and predefined inclusion criteria. The methodology of systematic reviews makes them reproducible which is not the case in narrative reviews [1,2,3]. The number of systematic reviews and meta-analyses on nuclear medicine diagnostic and prognostic studies is increasing [4, 5]. In the current chapter, a practical guideline has been prepared for the researchers who intend to perform a systematic review or meta-analysis of diagnostic and prognostic studies.
The single most important step in preparing a systematic review is to have a clear topic. The topic is usually divided into several aspects including: patients (the population of the study), intervention (the diagnostic test under study or a prognostic factor which is being evaluated), comparison (the procedures comparative to the index test), outcome (the outcome which is going to be evaluated which are usually sensitivity and specificity for diagnostic studies and overall survival (OR) and progression free survival (PFS) in prognostic ones).
The abovementioned method is called patients-intervention-comparison-outcome (PICO) [6, 7]. The search strategy for systematic reviews is based on the PICO question.
Here are two examples:
Search strategy is based on our PICO question. The keywords and databases which are used for searching should minimize the chance of missing any relevant article. Using Boolean operators (i.e., AND, OR, NOT) is highly recommended. This makes your search as sensitive as possible.
For example for the abovementioned PICO questions the following keywords seem to be optimal:
For example for the abovementioned PICO questions, the following inclusion criteria can be set:
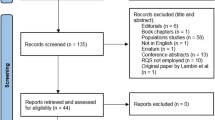
Full texts of all relevant studies should be retrieved. The reference of primary studies and all relevant reviews should be checked to search for additional primary studies that could have been missed (backward searching of the citations). In addition, articles citing the relevant included articles can be used to find any other missing articles (forward searching of the citations). The citing articles can be found easily using Google Scholar (https://scholar.google.com/), SCOPUS, or Dimensions (a free newly launched application with many useful options: https://app.dimensions.ai/discover/publication).
Remember to keep the records of all the searches, as well as included and excluded studies.
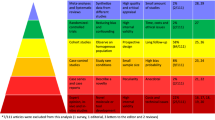
Not all included studies are of same quality. Quality of each study should be checked and reported. Several checklists are available for diagnostic studies [8, 9].
Two of the most commonly used checklists are:
Several checklists are also available for prognostic studies [11].
Two of the most commonly used checklists are:
Checklists are usually equivalent to each other; however, each domain or dimension for all included studies should be explained in detail to give the reader of systematic reviews a clue regarding the quality of the included studies. Reporting only based on numbers (quality scores) should be discouraged.
All relevant data should be extracted from the included studies. Detailed information regarding the study population, method of the diagnostic or prognostic test, gold standard test, follow-up times, methods of ascertaining outcomes, outcome variables such as false and true negative (FN, TN), false and true positive (FP, TP) cases for diagnostic studies, and hazard ratios (HR) for OS and PFS for prognostic studies should be extracted. Extraction of data should be as complete as possible to allow reconstruction of 2 × 2 diagnostic tables or HR of prognostic factors as well as sub-group analyses [11, 13, 14].
Extraction of data in prognostic studies can be very tricky: not all studies reported HR, and only Kaplan Meier (KM) curves and associated log rank tests are usually reported. HR can be extracted from KM curves. Usually the survival data can be extracted manually from KM curves using special software such as getdata graph digitizer (available at http://getdata-graph-digitizer.com/download.php). Finally the extracted survival data can be converted to HR by Parmar method using a special Excel file provided by Parmar et al. [15].
Another important aspect of extraction data in prognostic systematic reviews is type of prognostic factor (quantitative vs. qualitative factors) and evaluation of other prognostic factors (multivariate vs. univariate analysis). HR of quantitative variables (such as SUVmax) can be provided in two ways: first, the prognostic factor can be used as a quantitative variable and a HR using Cox regression is provided. The second type of HR can be calculated by categorizing a quantitative variable into two ranks (for example, SUVmax >7 and ≤7). These two types of HR cannot be pooled with each other even for a same prognostic factor. In addition, only univariate or multivariate HR should be used for pooling data across studies. Pooling univariate HR with a multivariate HR is discouraged as the latter (but not the former) takes into account other potential prognostic factors.
In this final step, the numerical results of the included studies would be pooled together. First of all, diagnostic or prognostic indices of each included study should be presented.
The following diagnostic indices should be reported:
The following prognostic indices should be reported:
Meta-analysis is a special statistical method for pooling data across different studies and giving pooled diagnostic indices. For this purpose, a weight is attributed to each study and the weighted diagnostic indices are pooled together. Special software are available for this purpose, including SAS, R, and STATA.
For diagnostic studies, two free software are available:
For prognostic studies, usually hazard ratios should be pooled across included studies. Several software are available in this regard, such as R, SAS, and Comprehensive Meta-Analysis (CMA).
The least required data to be provided in a meta-analysis are:
The discussion and final conclusion of a systematic review and meta-analysis should be as objective as possible. The authors should discuss the main results of the systematic review and meta-analysis. Final conclusion should be based on the main results of the systematic review. Any heterogeneity of the included studies should be explained and the possible reasons should be discussed.
Standard method of reporting systematic reviews and meta-analyses Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) provides a minimum requirement for reporting systematic reviews and meta-analyses [21, 26]. Although it is originally prepared for systematic reviews of randomized clinical trials, systematic reviews of diagnostic accuracy studies can be reported using PRISMA too. PRISMA statement and checklist can be found in the following link: http://www.prisma-statement.org/.
To publish a high quality systematic review or meta-analysis of diagnostic test accuracy or prognostic studies, certain methodology should be followed. Only methodologically sound systematic reviews and meta-analyses are worth publication and can change or support clinical use of a diagnostic test or a prognostic factor. Hopefully, the abovementioned methodology could help the researchers through the process of systematic review and meta-analysis preparation.